Profiling
Quick profiling in your terminal
Note
This is only meant to be used for quick profiling or programmatically accessing the profiling results. For more detailed and GUI friendly profiling proceed to the next section.
Simply replace the use of Base.@time or Base.@timed with Reactant.Profiler.@time or Reactant.Profiler.@timed. We will automatically compile the function if it is not already a Reactant compiled function (with sync=true).
using Reactant
x = Reactant.to_rarray(randn(Float32, 100, 2))
W = Reactant.to_rarray(randn(Float32, 10, 100))
b = Reactant.to_rarray(randn(Float32, 10))
linear(x, W, b) = (W * x) .+ b
Reactant.@time linear(x, W, b)WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
I0000 00:00:1771371395.019282 4306 profiler_session.cc:117] Profiler session initializing.
I0000 00:00:1771371395.019318 4306 profiler_session.cc:132] Profiler session started.
I0000 00:00:1771371395.019626 4306 profiler_session.cc:81] Profiler session collecting data.
I0000 00:00:1771371395.020192 4306 save_profile.cc:150] Collecting XSpace to repository: /tmp/reactant_profile/plugins/profile/2026_02_17_23_36_35/runnervmjduv7.xplane.pb
I0000 00:00:1771371395.020340 4306 save_profile.cc:123] Creating directory: /tmp/reactant_profile/plugins/profile/2026_02_17_23_36_35
I0000 00:00:1771371395.020452 4306 save_profile.cc:129] Dumped gzipped tool data for trace.json.gz to /tmp/reactant_profile/plugins/profile/2026_02_17_23_36_35/runnervmjduv7.trace.json.gz
I0000 00:00:1771371395.020469 4306 profiler_session.cc:150] Profiler session tear down.
I0000 00:00:1771371395.034536 4306 stub_factory.cc:163] Created gRPC channel for address: 0.0.0.0:40067
I0000 00:00:1771371395.034837 4306 grpc_server.cc:94] Server listening on 0.0.0.0:40067 with max_concurrent_requests 1
I0000 00:00:1771371395.034870 4306 xplane_to_tools_data_with_profile_processor.cc:141] serving tool: memory_profile with options: {} using ProfileProcessor
I0000 00:00:1771371395.034879 4306 xplane_to_tools_data_with_profile_processor.cc:163] Using local processing for tool: memory_profile
I0000 00:00:1771371395.034883 4306 memory_profile_processor.cc:47] Processing memory profile for host: runnervmjduv7
I0000 00:00:1771371395.035218 4306 xplane_to_tools_data_with_profile_processor.cc:168] Total time for tool memory_profile: 345.015us
I0000 00:00:1771371395.048107 4306 xplane_to_tools_data_with_profile_processor.cc:141] serving tool: op_profile with options: {} using ProfileProcessor
I0000 00:00:1771371395.048137 4306 xplane_to_tools_data_with_profile_processor.cc:163] Using local processing for tool: op_profile
I0000 00:00:1771371395.048288 4306 xprof_thread_pool_executor.cc:22] Creating derived_timeline_trace_events XprofThreadPoolExecutor with 4 threads.
I0000 00:00:1771371395.049118 4306 xprof_thread_pool_executor.cc:22] Creating ProcessTensorCorePlanes XprofThreadPoolExecutor with 4 threads.
I0000 00:00:1771371395.052308 4306 xprof_thread_pool_executor.cc:22] Creating op_stats_threads XprofThreadPoolExecutor with 4 threads.
I0000 00:00:1771371395.053244 4306 xplane_to_tools_data_with_profile_processor.cc:168] Total time for tool op_profile: 5.130421ms
I0000 00:00:1771371395.062291 4306 xplane_to_tools_data_with_profile_processor.cc:141] serving tool: overview_page with options: {} using ProfileProcessor
I0000 00:00:1771371395.062303 4306 xplane_to_tools_data_with_profile_processor.cc:163] Using local processing for tool: overview_page
I0000 00:00:1771371395.062307 4306 overview_page_processor.cc:64] OverviewPageProcessor::ProcessSession
I0000 00:00:1771371395.062621 4306 xprof_thread_pool_executor.cc:22] Creating ConvertMultiXSpaceToInferenceStats XprofThreadPoolExecutor with 1 threads.
I0000 00:00:1771371395.063184 4306 xplane_to_tools_data_with_profile_processor.cc:168] Total time for tool overview_page: 885.385us
runtime: 0.00025467s
compile time: 4.67539459sReactant.@timed nrepeat=100 linear(x, W, b)AggregateProfilingResult(
runtime = 0.00025467s,
compile_time = 0.13004362s, )Note that the information returned depends on the backend. Specifically CUDA and TPU backends provide more detailed information regarding memory usage and allocation (something like the following will be displayed on GPUs):
AggregateProfilingResult(
runtime = 0.00003829s,
compile_time = 2.18053260s, # time spent compiling by Reactant
GPU_0_bfc = MemoryProfileSummary(
peak_bytes_usage_lifetime = 64.010 MiB, # peak memory usage over the entire program (lifetime of memory allocator)
peak_stats = MemoryAggregationStats(
stack_reserved_bytes = 0 bytes, # memory usage by stack reservation
heap_allocated_bytes = 30.750 KiB, # memory usage by heap allocation
free_memory_bytes = 23.518 GiB, # free memory available for allocation or reservation
fragmentation = 0.514931, # fragmentation of memory within [0, 1]
peak_bytes_in_use = 30.750 KiB # The peak memory usage over the entire program
)
peak_stats_time = 0.04975365s,
memory_capacity = 23.518 GiB # memory capacity of the allocator
)
flops = FlopsSummary(
Flops = 2.8369974648038653e-9, # [flops / (peak flops * program time)], capped at 1.0
UncappedFlops = 2.8369974648038653e-9,
RawFlops = 4060.0, # Total FLOPs performed
BF16Flops = 4060.0, # Total FLOPs Normalized to the bf16 (default) devices peak bandwidth
RawTime = 0.00040298422s, # Raw time in seconds
RawFlopsRate = 1.0074836180930361e7, # Raw FLOPs rate in FLOPs/seconds
BF16FlopsRate = 1.0074836180930361e7, # BF16 FLOPs rate in FLOPs/seconds
)
)Additionally for GPUs and TPUs, we can use the Reactant.@profile macro to profile the function and get information regarding each of the kernels executed.
Reactant.@profile linear(x, W, b)I0000 00:00:1771371395.747435 4306 profiler_session.cc:117] Profiler session initializing.
I0000 00:00:1771371395.748279 4306 profiler_session.cc:132] Profiler session started.
I0000 00:00:1771371395.748445 4306 profiler_session.cc:81] Profiler session collecting data.
I0000 00:00:1771371395.748786 4306 save_profile.cc:150] Collecting XSpace to repository: /tmp/reactant_profile/plugins/profile/2026_02_17_23_36_35/runnervmjduv7.xplane.pb
I0000 00:00:1771371395.749016 4306 save_profile.cc:123] Creating directory: /tmp/reactant_profile/plugins/profile/2026_02_17_23_36_35
I0000 00:00:1771371395.749185 4306 save_profile.cc:129] Dumped gzipped tool data for trace.json.gz to /tmp/reactant_profile/plugins/profile/2026_02_17_23_36_35/runnervmjduv7.trace.json.gz
I0000 00:00:1771371395.749207 4306 profiler_session.cc:150] Profiler session tear down.
I0000 00:00:1771371395.749278 4306 xplane_to_tools_data_with_profile_processor.cc:141] serving tool: memory_profile with options: {} using ProfileProcessor
I0000 00:00:1771371395.749284 4306 xplane_to_tools_data_with_profile_processor.cc:163] Using local processing for tool: memory_profile
I0000 00:00:1771371395.749286 4306 memory_profile_processor.cc:47] Processing memory profile for host: runnervmjduv7
I0000 00:00:1771371395.749437 4306 xplane_to_tools_data_with_profile_processor.cc:168] Total time for tool memory_profile: 158.016us
I0000 00:00:1771371395.749461 4306 xplane_to_tools_data_with_profile_processor.cc:141] serving tool: op_profile with options: {} using ProfileProcessor
I0000 00:00:1771371395.749466 4306 xplane_to_tools_data_with_profile_processor.cc:163] Using local processing for tool: op_profile
I0000 00:00:1771371395.749552 4306 xplane_to_tools_data_with_profile_processor.cc:168] Total time for tool op_profile: 89.046us
I0000 00:00:1771371395.749572 4306 xplane_to_tools_data_with_profile_processor.cc:141] serving tool: overview_page with options: {} using ProfileProcessor
I0000 00:00:1771371395.749575 4306 xplane_to_tools_data_with_profile_processor.cc:163] Using local processing for tool: overview_page
I0000 00:00:1771371395.749577 4306 overview_page_processor.cc:64] OverviewPageProcessor::ProcessSession
I0000 00:00:1771371395.749670 4306 xprof_thread_pool_executor.cc:22] Creating ConvertMultiXSpaceToInferenceStats XprofThreadPoolExecutor with 1 threads.
I0000 00:00:1771371395.750243 4306 xplane_to_tools_data_with_profile_processor.cc:168] Total time for tool overview_page: 672.317us
I0000 00:00:1771371395.845184 4306 xplane_to_tools_data_with_profile_processor.cc:141] serving tool: kernel_stats with options: {} using ProfileProcessor
I0000 00:00:1771371395.845214 4306 xplane_to_tools_data_with_profile_processor.cc:163] Using local processing for tool: kernel_stats
I0000 00:00:1771371395.845369 4306 xplane_to_tools_data_with_profile_processor.cc:168] Total time for tool kernel_stats: 160.56us
I0000 00:00:1771371395.960457 4306 xplane_to_tools_data_with_profile_processor.cc:141] serving tool: framework_op_stats with options: {} using ProfileProcessor
I0000 00:00:1771371395.960486 4306 xplane_to_tools_data_with_profile_processor.cc:163] Using local processing for tool: framework_op_stats
I0000 00:00:1771371395.960724 4306 xplane_to_tools_data_with_profile_processor.cc:168] Total time for tool framework_op_stats: 242.484us
╔================================================================================╗
║ SUMMARY ║
╚================================================================================╝
AggregateProfilingResult(
runtime = 0.00025467s,
compile_time = 0.12748215s, # time spent compiling by Reactant
)On GPUs this would look something like the following:
╔================================================================================╗
║ KERNEL STATISTICS ║
╚================================================================================╝
┌───────────────────┬─────────────┬────────────────┬──────────────┬──────────────┬──────────────┬──────────────┬───────────┬──────────┬────────────┬─────────────┐
│ Kernel Name │ Occurrences │ Total Duration │ Avg Duration │ Min Duration │ Max Duration │ Static Shmem │ Block Dim │ Grid Dim │ TensorCore │ Occupancy % │
├───────────────────┼─────────────┼────────────────┼──────────────┼──────────────┼──────────────┼──────────────┼───────────┼──────────┼────────────┼─────────────┤
│ gemm_fusion_dot_1 │ 1 │ 0.00000250s │ 0.00000250s │ 0.00000250s │ 0.00000250s │ 2.000 KiB │ 64,1,1 │ 1,1,1 │ ✗ │ 100.0% │
│ loop_add_fusion │ 1 │ 0.00000131s │ 0.00000131s │ 0.00000131s │ 0.00000131s │ 0 bytes │ 20,1,1 │ 1,1,1 │ ✗ │ 31.2% │
└───────────────────┴─────────────┴────────────────┴──────────────┴──────────────┴──────────────┴──────────────┴───────────┴──────────┴────────────┴─────────────┘
╔================================================================================╗
║ FRAMEWORK OP STATISTICS ║
╚================================================================================╝
┌───────────────────┬─────────┬─────────────┬─────────────┬─────────────────┬───────────────┬──────────┬───────────┬──────────────┬──────────┐
│ Operation │ Type │ Host/Device │ Occurrences │ Total Self-Time │ Avg Self-Time │ Device % │ Memory BW │ FLOP Rate │ Bound By │
├───────────────────┼─────────┼─────────────┼─────────────┼─────────────────┼───────────────┼──────────┼───────────┼──────────────┼──────────┤
│ gemm_fusion_dot.1 │ Unknown │ Device │ 1 │ 0.00000250s │ 0.00000250s │ 65.55% │ 1.82 GB/s │ 1.6 GFLOP/s │ HBM │
│ +/add │ add │ Device │ 1 │ 0.00000131s │ 0.00000131s │ 34.45% │ 0.14 GB/s │ 0.05 GFLOP/s │ HBM │
└───────────────────┴─────────┴─────────────┴─────────────┴─────────────────┴───────────────┴──────────┴───────────┴──────────────┴──────────┘
╔================================================================================╗
║ SUMMARY ║
╚================================================================================╝
AggregateProfilingResult(
runtime = 0.00005622s,
compile_time = 2.32802137s, # time spent compiling by Reactant
GPU_0_bfc = MemoryProfileSummary(
peak_bytes_usage_lifetime = 64.010 MiB, # peak memory usage over the entire program (lifetime of memory allocator)
peak_stats = MemoryAggregationStats(
stack_reserved_bytes = 0 bytes, # memory usage by stack reservation
heap_allocated_bytes = 81.750 KiB, # memory usage by heap allocation
free_memory_bytes = 23.518 GiB, # free memory available for allocation or reservation
fragmentation = 0.514564, # fragmentation of memory within [0, 1]
peak_bytes_in_use = 81.750 KiB # The peak memory usage over the entire program
)
peak_stats_time = 0.00608052s,
memory_capacity = 23.518 GiB # memory capacity of the allocator
)
flops = FlopsSummary(
Flops = 2.033375207640664e-8, # [flops / (peak flops * program time)], capped at 1.0
UncappedFlops = 2.033375207640664e-8,
RawFlops = 4060.0, # Total FLOPs performed
BF16Flops = 4060.0, # Total FLOPs Normalized to the bf16 (default) devices peak bandwidth
RawTime = 0.00005622s, # Raw time in seconds
RawFlopsRate = 7.220987105380169e7, # Raw FLOPs rate in FLOPs/seconds
BF16FlopsRate = 7.220987105380169e7, # BF16 FLOPs rate in FLOPs/seconds
)
)Capturing traces
When running Reactant, it is possible to capture traces using the XLA profiler. These traces can provide information about where the XLA specific parts of program spend time during compilation or execution. Note that tracing and compilation happen on the CPU even though the final execution is aimed to run on another device such as GPU or TPU. Therefore, including tracing and compilation in a trace will create annotations on the CPU.
Let's setup a simple function which we can then profile
using Reactant
x = Reactant.to_rarray(randn(Float32, 100, 2))
W = Reactant.to_rarray(randn(Float32, 10, 100))
b = Reactant.to_rarray(randn(Float32, 10))
linear(x, W, b) = (W * x) .+ blinear (generic function with 1 method)The profiler can be accessed using the Reactant.with_profiler function.
Reactant.with_profiler("./") do
mylinear = Reactant.@compile linear(x, W, b)
mylinear(x, W, b)
end10×2 ConcretePJRTArray{Float32,2}:
7.34116 3.7869
-7.14728 -16.7097
16.5525 -4.75952
-11.5283 -1.36271
-13.534 14.1503
-10.1889 2.60556
3.62489 -0.252602
-1.04012 -0.447452
-8.46486 -0.523882
-5.49211 -0.0569211Running this function should create a folder called plugins in the folder provided to Reactant.with_profiler which will contain the trace files. The traces can then be visualized in different ways.
Note
For more insights about the current state of Reactant, it is possible to fetch device information about allocations using the Reactant.XLA.allocatorstats function.
Perfetto UI
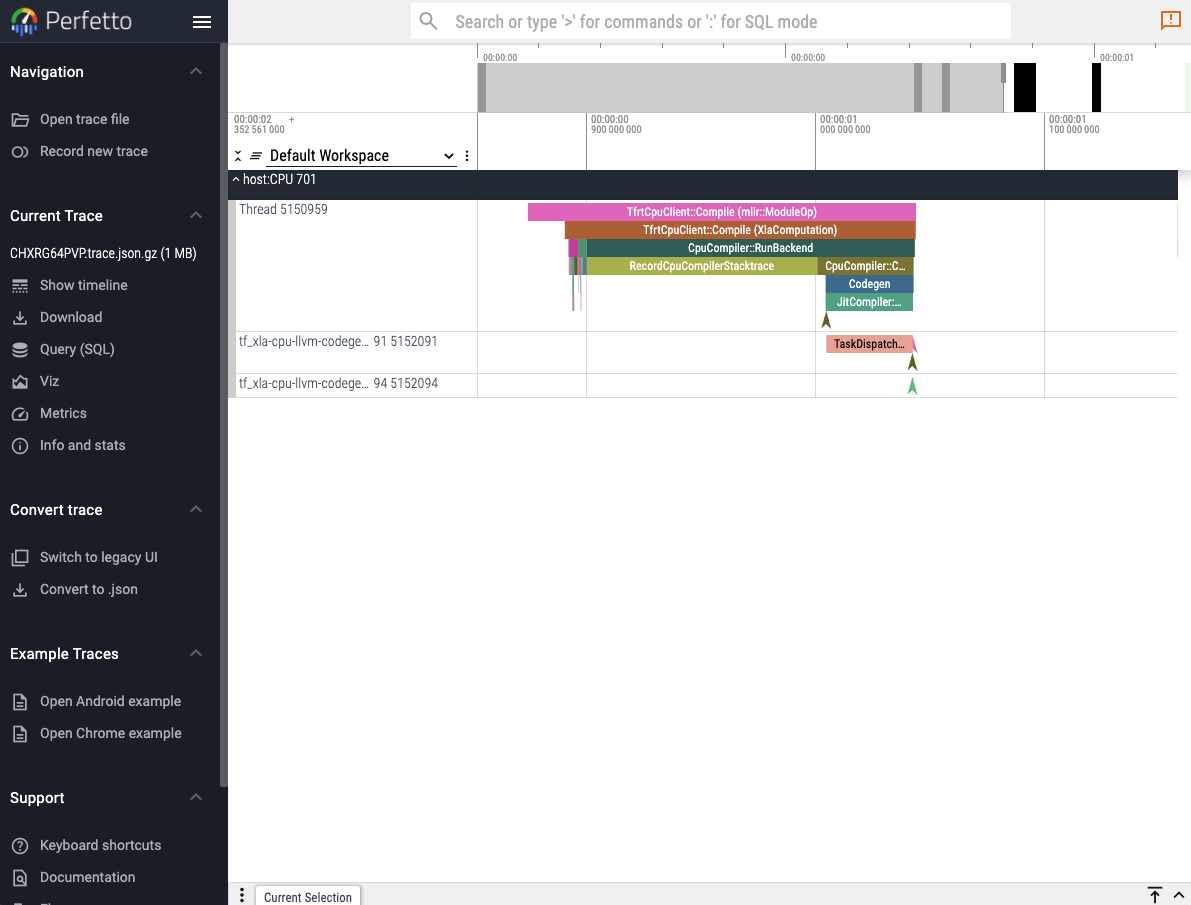
The first and easiest way to visualize a captured trace is to use the online perfetto.dev tool. Reactant.with_profiler has a keyword parameter called create_perfetto_link which will create a usable perfetto URL for the generated trace. The function will block execution until the URL has been clicked and the trace is visualized. The URL only works once.
Reactant.with_profiler("./"; create_perfetto_link=true) do
mylinear = Reactant.@compile linear(x, W, b)
mylinear(x, W, b)
endNote
It is recommended to use the Chrome browser to open the perfetto URL.
Tensorboard
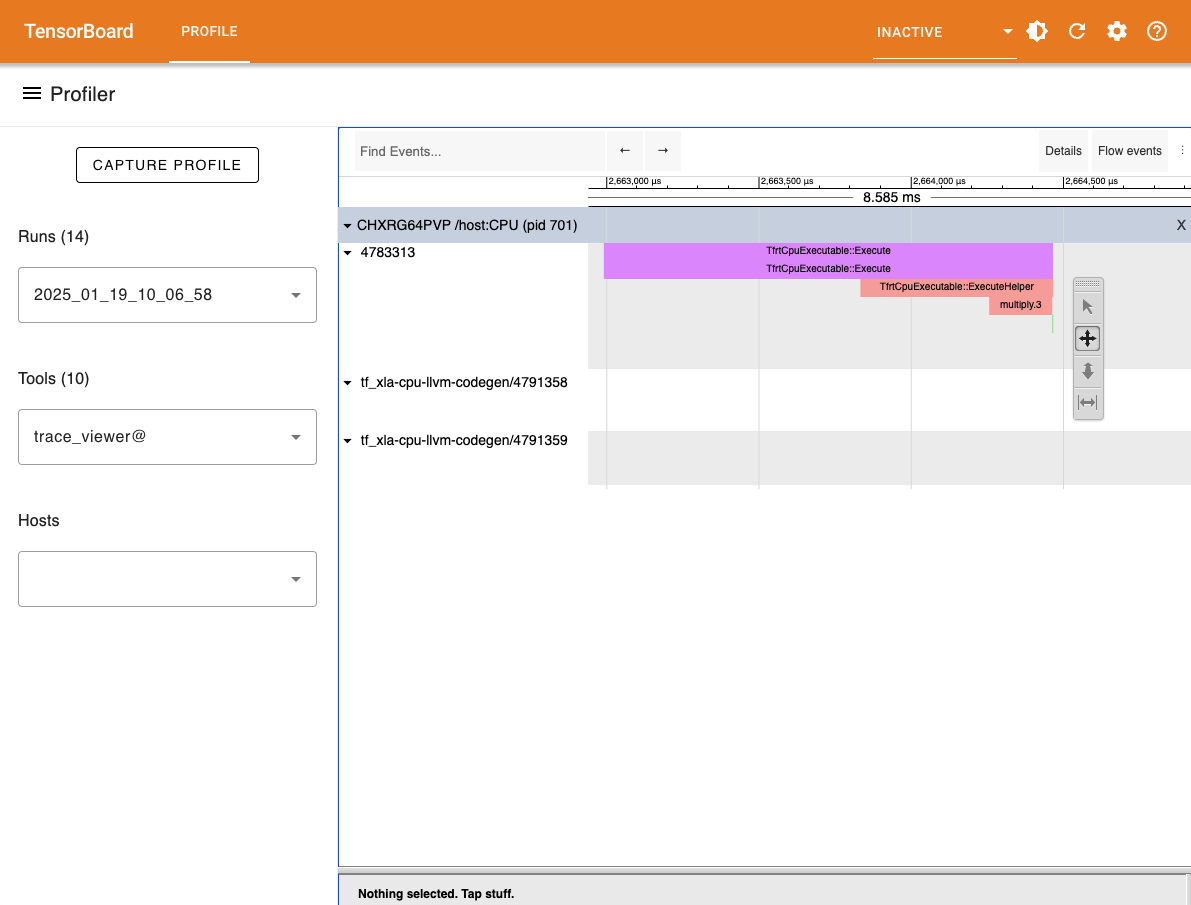
Another option to visualize the generated trace files is to use the tensorboard profiler plugin. The tensorboard viewer can offer more details than the timeline view such as visualization for compute graphs.
First install tensorboard and its profiler plugin:
pip install tensorboard tensorboard-plugin-profileAnd then run the following in the folder where the plugins folder was generated:
tensorboard --logdir ./Adding Custom Annotations
By default, the traces contain only information captured from within XLA. The Reactant.Profiler.annotate function can be used to annotate traces for Julia code evaluated during tracing.
Reactant.Profiler.annotate("my_annotation") do
# Do things...
endThe added annotations will be captured in the traces and can be seen in the different viewers along with the default XLA annotations. When the profiler is not activated, then the custom annotations have no effect and can therefore always be activated.